Predicting the potential for zoonotic transmission and host associations for novel viruses
Pandit PS, Anthony SJ, Goldstein T, Olival KJ, Doyle MM, Gardner NR, Bird B, Smith WA, Wolking D, Gilardi K, Monagin C, Kelly T, Uhart M, Epstein JH, Machalaba C, Rostal MK, Dawson P, Hagan E, Sullivan A, Li H, Chmura AA, Latinne A, Lange C, O’Rourke T, Olson SH, Keatts L, Mendoza AP, Perez A, de Paula CD, Zimmerman D, Valitutto M, LeBreton M, McIver D, Islam A, Duong V, Mouiche M, Shi Z, Mulembakani P, Kumakamba C, Ali M, Kebede N, Tamoufe U, Bel-Nono S, Camara A, Pamungkas J, Coulibaly K, Abu-Basha E, Kamau J, Silithammavong S, Desmond J, Hughes T, Shiilegdamba E, Aung O, Karmacharya D, Nziza J, Ndiaye D, Gbakima A, Sijali Z, Wacharapluesadee S, Robles EA, Ssebide B, Suzán G, Aguirre LF, Solorio MR, Dhole TN, Nga NTT, Hitchens PL, Joly DO, Saylors K, Fine A, Murray S, Karesh W, Daszak P, Mazet JAK, PREDICT Consortium, de Paula CD, Johnson CK. Communications Biology. 2022 Aug 19; 5:844. https://doi.org/10.1038/s42003-022-03797-9
Our virus-host association network predicts spillover risk and potential hosts for novel viruses detected in wildlife
This research predicts zoonotic transmission risk and potential hosts for 531 novel animal viruses, primarily of wildlife origin, based on virus-host associations of known viruses. Improving our understanding of the ecological characteristics of novel viruses will allow us to better assess which newly discovered viruses hold the potential to spread to humans, domestic species, or other wildlife. Results from studies like this can inform on the ecology of novel viruses and help prioritize viruses for further in-vitro or in-vivo characterization.
Jump to: Key Findings | Figures | How Our Models Work | Significance | Next Steps
Key Findings
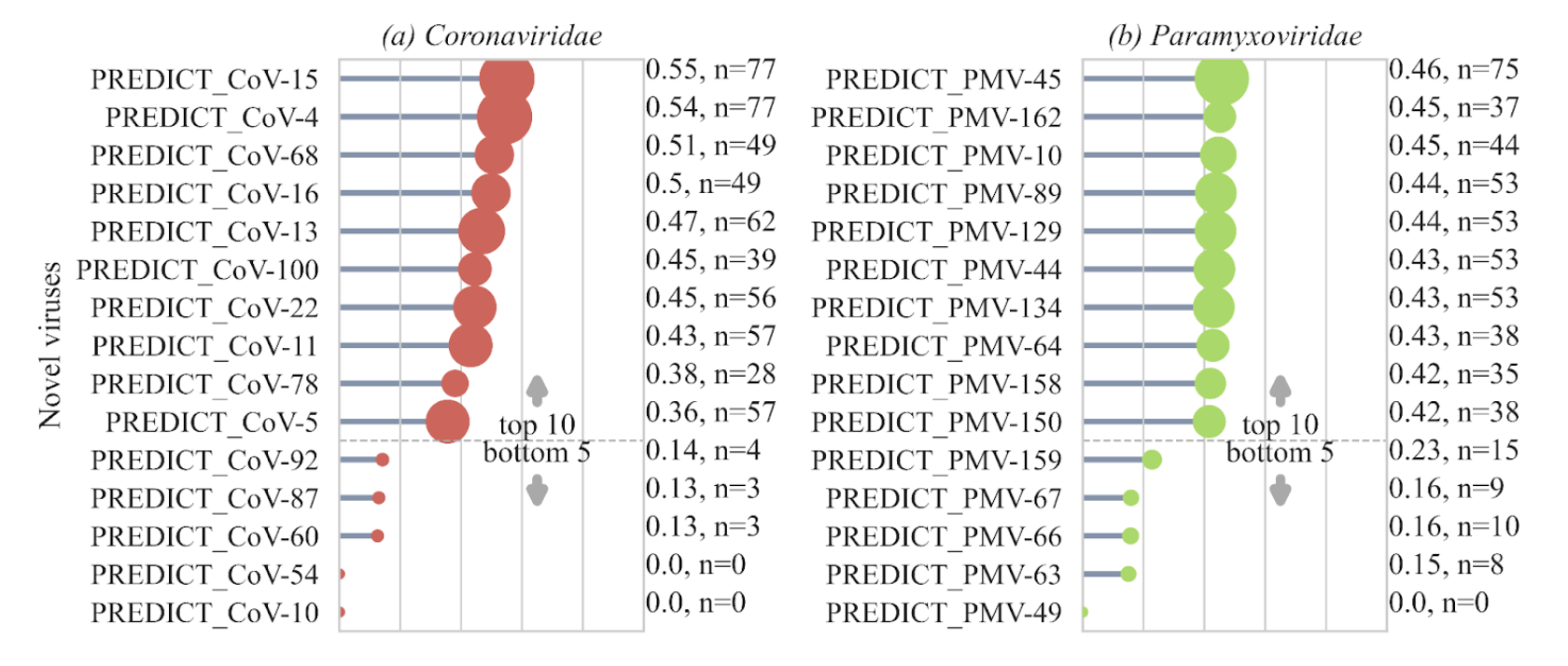
- Prioritization scores generated for 531 novel wildlife viruses detected around the world allow us to better assess zoonotic transmission risk.
- ❯ The prioritization score provides a data driven tool to quantify the ecological and evolutionary trajectory towards zoonotic transmission for novel viruses, with higher scores indicating greater risk.
❯ Coronaviruses with high prioritization scores were detected in various bat species from the Phyllostomidae, Hipposideridae, Vespertilionidae, and Pteropodidae families. More surveillance efforts are needed for bat species found in South America and Southeast Asia within these families.
❯ PREDICT_CoV-78, which was detected in bats and rodents in Southeast Asia, also showed a high prioritization score. This is a rare detection of a novel virus shared across different taxonomic orders
- Novel coronaviruses are predicted to have greater host plasticity compared to novel viruses from other families.
- ❯ Novel coronaviruses are predicted to have higher host plasticity than novel viruses from other viral families, suggesting they are more likely to be found in multiple different animal species than other viruses.
❯ This is indicated by a higher predicted network degree and betweenness centrality for those viruses, the network metric assessing connections.
- Novel viruses detected during a decade of recent wildlife surveillance are more host specific than well recognized viruses.
- ❯ This is indicated through lower network centrality distributions of predicted host-virus network, the host-virus network generated after including prediction for novel viruses.
❯ Zoonotic viruses are known to have wider host breath than non-zoonotic viruses. Having an estimation on the host breath helps characterize the zoonotic risk of a novel or under-researched virus.
Figures
Modeling workflow
How Our Models Work
Determining the potential for zoonotic transmission risk for newly discovered viruses is challenging and time-consuming, especially with the limited data available for any novel virus. To accomplish this, we developed machine learning models that use network characteristics to predict the connections between these viruses, i.e. nodes of the network, and also predict which taxonomic groups will share viruses, including if viruses are shared with humans. These models were trained on a network of vertebrate hosts of viruses that humans and animals share (zoonotic) and animal-only viruses (non-zoonotic) to develop a data-driven prioritization scoring system. The predictive abilities of the models gave us insights into potential host range that we can estimate for novel viruses to understand the zoonotic risk and host preference.
While the model is trained on numerous data points collected from decades of virus transmission research available from online repositories and published literature on well-recognized zoonotic and non-zoonotic animal viruses, the model only needs the virus family and the host in which the virus was found to generate predictions for new viruses. Both of these data points are known for most newly discovered viruses, which allows our model to make predictions about the potential for the new virus to infect humans, and helps inform if further study of the virus is warranted. We used our model to generate predictions on novel viruses detected in wildlife over ten years of surveillance sampling conducted in over 30 countries throughout Africa, Asia, and Latin America. Model results highlight key ecological characteristics of novel viruses, such as zoonotic transmission risk and potential hosts.
Data and code from these analyses are available at:
Significance
Previous studies have tried understanding the risk based on expert opinions. Our approach develops a more agnostic and data-driven metric to understand the risk of zoonotic spillover and prioritize viruses accordingly for further in-vitro and in-vivo characterization. With additional data and the inclusion of other predictive features, including molecular characteristics, these models will improve significantly in their prediction ability. The prioritization of novel viruses for further characterization, and increased availability of ecological traits and genomic data resulting from these broad surveillance efforts will also provide improved model predictions.
Next Steps
Given that our findings provide further evidence for the relationship between higher host plasticity and greater zoonotic potential, key future research directions would involve additional surveillance across a broad taxonomic range to gain insights on newly detected viruses to further inform on spillover risk.
As virus discovery surveillance programs explore the virome, we will be soon inundated with virus data. Understanding the risk from all these new findings will be crucial to ensure that future pandemics are prevented. Tools and approaches like these will pave the way to streamline our understanding of the risks these viruses pose to human and wildlife health.
Some of our current projects are exploring the relationships between landscape change, virus transmission, and spillover. Learn more about them here:
EpiCenter for Emerging Infectious Disease Intelligence Pathogen Plasticity Project
This work was supported by the United States Agency for International Development Emerging Pandemic Threat PREDICT program (Cooperative Agreements GHN-A-00-09-00010-00 and AID-OAA-A-14-00102) and by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (award U01AI151814). The research shared here is solely the responsibility of the authors and does not necessarily represent the official views of the USAID, National Institutes of Health, or the United States Government.